AI and Machine Learning in Single-Cell Transcriptomic Analysis
Introduction
In recent years, single-cell transcriptomics has revolutionized our understanding of biology by enabling researchers to explore gene expression at the resolution of individual cells. This breakthrough technology provides unprecedented insight into the diversity of cell types, developmental processes, and disease mechanisms. However, the complexity and scale of single-cell transcriptomic data present significant analytical challenges. Enter artificial intelligence (AI) and machine learning (ML) - powerful tools that are transforming how scientists interpret and gain insights from these data-rich experiments.
Definition
Single-cell analysis transcriptomics is a technique used to study gene expression at the level of individual cells. Unlike traditional bulk RNA sequencing, which measures average gene activity across many cells, this method captures the unique transcriptomic profile of each cell, revealing cellular heterogeneity and enabling the identification of rare cell types, developmental pathways, and disease mechanisms.
Understanding Single-Cell Transcriptomics
Single-cell transcriptomics involves isolating individual cells and sequencing their RNA to capture the expression levels of thousands of genes per cell. Unlike bulk RNA sequencing, which provides an average expression profile across many cells, single-cell RNA sequencing (scRNA-seq) can detect subtle differences between individual cells, uncovering rare cell populations and revealing cellular heterogeneity in tissues.
This data-rich approach is widely applied in developmental biology, immunology, cancer research, and neuroscience. Yet, scRNA-seq datasets are often high-dimensional, sparse, and noisy, making them ideal candidates for advanced computational techniques like AI and ML.
The Role of AI and ML in Analyzing scRNA-seq Data
AI and ML methods have emerged as indispensable tools for overcoming the analytical challenges posed by single-cell data. These technologies can automatically detect patterns, cluster cell types, infer gene regulatory networks, and even predict cell fate transitions with minimal human intervention. Here are some of the most impactful applications:
Cell Type Identification and Clustering:
One of the first steps in scRNA-seq analysis is grouping similar cells based on their gene expression profiles. Traditionally, this has been done using unsupervised clustering algorithms such as k-means or hierarchical clustering. However, machine learning methods like t-SNE (t-distributed stochastic neighbor embedding), UMAP (Uniform Manifold Approximation and Projection), and more recently, autoencoders and deep learning-based approaches, have significantly improved the visualization and resolution of cell clusters.
Deep learning models can capture complex, nonlinear relationships in high-dimensional data and are increasingly being used for dimensionality reduction and clustering. For instance, Variational Autoencoders (VAEs) and Generative Adversarial Networks (GANs) have been applied to generate more informative low-dimensional representations of transcriptomic data.
Data Imputation and Noise Reduction:
scRNA-seq data is often noisy and contains dropout events — where the expression of a gene is recorded as zero due to technical limitations rather than biological absence. Machine learning algorithms can help impute missing values and denoise data to improve downstream analyses.
DeepImpute, MAGIC, and scImpute are some of the ML-based tools developed for this purpose. These models use information from similar cells to predict and fill in missing expression values, enabling more accurate analysis of gene expression dynamics.
Trajectory Inference and Pseudotime Analysis:
In developmental biology, it's crucial to understand how cells transition from one state to another. Machine learning techniques can infer trajectories — or “pseudotime” — of cell development, providing a temporal ordering of cells based on gene expression changes.
Tools like Monocle, Slingshot, and PAGA use various ML methods to model dynamic biological processes. More advanced models like neural ordinary differential equations (neural ODEs) have also been explored for capturing the continuous evolution of cell states.
Gene Regulatory Network Inference:
Understanding how genes interact to control cell behavior is central to functional genomics. AI-driven models can infer gene regulatory networks (GRNs) from single-cell data by learning patterns of gene co-expression and causality.
ML techniques such as Random Forests, Bayesian networks, and deep learning frameworks are commonly employed. Tools like SCENIC (Single-Cell Regulatory Network Inference and Clustering) integrate machine learning to identify active transcription factors and their target genes in different cell states.
Cell Classification and Annotation:
AI and ML algorithms can be trained to classify cells into predefined types based on labeled datasets. This is especially valuable for large-scale projects like the Human Cell Atlas, where rapid and accurate cell annotation is essential.
Supervised learning models — including support vector machines (SVMs), neural networks, and ensemble methods — can generalize from training data to classify new cells with high accuracy. These models are increasingly integrated into automated pipelines for single-cell data analysis.
Integration of Multi-Omics Data:
Single-cell studies often go beyond transcriptomics, incorporating other layers like epigenomics (scATAC-seq), proteomics, and spatial transcriptomics. Integrating these data types is a complex challenge, but AI models excel at learning from diverse and multimodal datasets.
Machine learning techniques such as multimodal autoencoders and canonical correlation analysis (CCA) help align and interpret data across different omics platforms. This integrative approach provides a more holistic view of cellular function and disease pathology.
Challenges and Considerations
While AI and ML hold great promise, there are also challenges that researchers must consider:
Interpretability: It might be challenging to comprehend how predictions are formed by deep learning models since they can be "black boxes." Efforts are underway to develop explainable AI methods in bioinformatics.
Data Quality and Labeling: Machine learning models rely heavily on the quality and quantity of training data. Poorly annotated or noisy datasets can lead to misleading results.
Computational Resources: Training complex models requires significant computational power and memory, which may limit accessibility for some research groups.
Reproducibility: Ensuring that results can be reproduced across different datasets, labs, and analysis pipelines remains an ongoing concern in the field.
The Future of AI in Single-Cell Genomics
The convergence of single-cell biology and artificial intelligence is rapidly advancing our understanding of complex biological systems. As new ML algorithms become more robust, interpretable, and accessible, their integration into single-cell workflows will become standard practice.
Future developments may include:
Real-time analysis of single-cell data during sequencing experiments.
More comprehensive models that integrate time, space, and multimodal data.
Cloud-based AI platforms to democratize access to powerful computational tools.
Moreover, large-scale public datasets combined with open-source AI tools will empower researchers worldwide to extract deeper biological insights without the need for extensive coding or statistical expertise.
Growth Rate of Single-Cell Analysis Transcriptomics Market
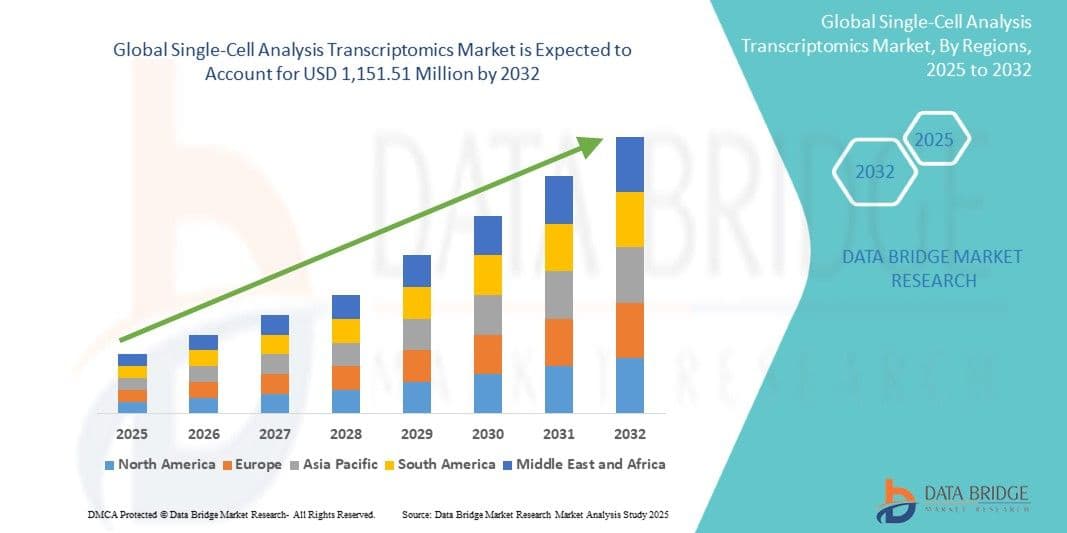
According to Data Bridge Market Research, the single-cell analysis transcriptomics market is projected to grow from its 2024 valuation of USD 633.07 million to USD 1,151.51 million by 2032. The market is expected to expand at a compound annual growth rate (CAGR) of 9.40% between 2025 and 2032, mainly due to the expected introduction of treatments that target single-cell gene expression.
Read More: https://www.databridgemarketresearch.com/reports/global-single-cell-analysis-transcriptomics-market
Conclusion
AI and machine learning are transforming the field of single-cell transcriptomic analysis by making it possible to process, visualize, and interpret complex biological data with greater accuracy and efficiency. From identifying cell types and inferring developmental trajectories to predicting gene regulatory networks, ML algorithms are becoming integral to the modern genomics toolkit. As both technologies continue to evolve, their synergy will undoubtedly unlock new discoveries in biology and medicine, bringing us closer to precision diagnostics, personalized therapies, and a deeper understanding of life at the cellular level.
Appreciate the creator